Basic Data Operations in R
Ani Ruhil
Basic data operations
- Common data types and modifications
- Case operations
- Subsetting data and factor levels
- Exporting data
- Merging data
- Dealing with duplicates
- Rearranging a data-frame
- Working with strings
Common data types
For the most part you'll focus on variables that are
- numeric -- the usual quantitative measures, also called "continuous" variables
- factors -- categorical variables such as sex, BMI categories, and so on
- dates -- but we'll deal with these in detail later
look at the variables in the hsb data
read.table('https://stats.idre.ucla.edu/stat/data/hsb2.csv', header = TRUE, sep = ",") -> hsbnames(hsb)## [1] "id" "female" "race" "ses" "schtyp" "prog" "read" "write" ## [9] "math" "science" "socst"str(hsb)## 'data.frame': 200 obs. of 11 variables:## $ id : int 70 121 86 141 172 113 50 11 84 48 ...## $ female : int 0 1 0 0 0 0 0 0 0 0 ...## $ race : int 4 4 4 4 4 4 3 1 4 3 ...## $ ses : int 1 2 3 3 2 2 2 2 2 2 ...## $ schtyp : int 1 1 1 1 1 1 1 1 1 1 ...## $ prog : int 1 3 1 3 2 2 1 2 1 2 ...## $ read : int 57 68 44 63 47 44 50 34 63 57 ...## $ write : int 52 59 33 44 52 52 59 46 57 55 ...## $ math : int 41 53 54 47 57 51 42 45 54 52 ...## $ science: int 47 63 58 53 53 63 53 39 58 50 ...## $ socst : int 57 61 31 56 61 61 61 36 51 51 ...Some of these variables are not really numeric, in particular:
- female = (0/1)
- race = (1=hispanic 2=asian 3=african-amer 4=white)
- ses = socioeconomic status (1=low 2=middle 3=high)
- schtyp = type of school (1=public 2=private)
- prog = type of program (1=general 2=academic 3=vocational)
We will have to convert these pseudo-numeric variables and label what the values represent
factor(hsb$female, levels = c(0, 1), labels = c("Male", "Female")) -> hsb$female factor(hsb$race, levels = c(1:4), labels = c("Hispanic", "Asian", "African American", "White")) -> hsb$race factor(hsb$ses, levels = c(1:3), labels = c("Low", "Middle", "High")) -> hsb$ses factor(hsb$schtyp, levels = c(1:2), labels = c("Public", "Private")) -> hsb$schtyp factor(hsb$prog, levels = c(1:3), labels = c("General", "Academic", "Vocational")) -> hsb$prog str(hsb)## 'data.frame': 200 obs. of 11 variables:## $ id : int 70 121 86 141 172 113 50 11 84 48 ...## $ female : Factor w/ 2 levels "Male","Female": 1 2 1 1 1 1 1 1 1 1 ...## $ race : Factor w/ 4 levels "Hispanic","Asian",..: 4 4 4 4 4 4 3 1 4 3 ...## $ ses : Factor w/ 3 levels "Low","Middle",..: 1 2 3 3 2 2 2 2 2 2 ...## $ schtyp : Factor w/ 2 levels "Public","Private": 1 1 1 1 1 1 1 1 1 1 ...## $ prog : Factor w/ 3 levels "General","Academic",..: 1 3 1 3 2 2 1 2 1 2 ...## $ read : int 57 68 44 63 47 44 50 34 63 57 ...## $ write : int 52 59 33 44 52 52 59 46 57 55 ...## $ math : int 41 53 54 47 57 51 42 45 54 52 ...## $ science: int 47 63 58 53 53 63 53 39 58 50 ...## $ socst : int 57 61 31 56 61 61 61 36 51 51 ...If we wanted to be prudent we could have created new factors, preserving the original data (my preference and now a habit)
read.table('https://stats.idre.ucla.edu/stat/data/hsb2.csv', header = TRUE, sep = ",") -> hsb factor(hsb$female, levels = c(0, 1), labels = c("Male", "Female")) -> hsb$female.f factor(hsb$race, levels = c(1:4), labels = c("Hispanic", "Asian", "African American", "White")) -> hsb$race.f factor(hsb$ses, levels = c(1:3), labels = c("Low", "Middle", "High")) -> hsb$ses.f factor(hsb$schtyp, levels = c(1:2), labels = c("Public", "Private")) -> hsb$schtyp.f factor(hsb$prog, levels = c(1:3), labels = c("General", "Academic", "Vocational")) -> hsb$prog.fAt times, we may have numbers that get read in as characters or factors, as is evident from the example below:
c(1, 2, 3, 4, 5) -> x1 c(1, 2, 3, 4, "a5") -> x2c(1, 2, 3, 4, "hello") -> x3c(1, 2, 3, 4, NA) -> x4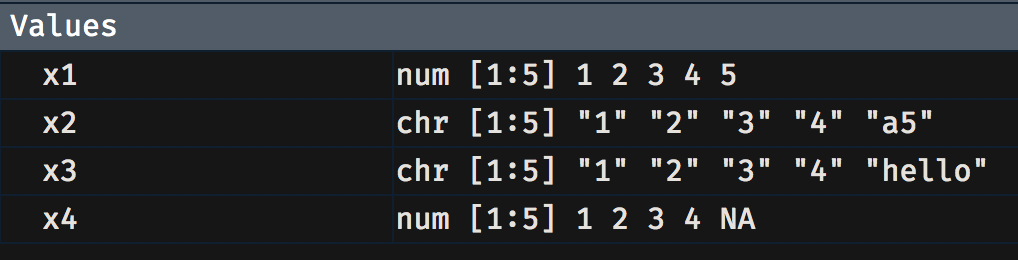
Notice that x1 and x2 are read in as numeric but x2 and x3 show up as chr, short for character. You could convert x2 and x3 into numeric as follows:
as.numeric(x2) -> x2.num as.numeric(x3) -> x3.numThe NA values are not applicable values, essentially missing values.
At times the variable may in fact be numeric but because of some anomalies in the variable, R will read it in as a factor. When this happens, converting the variable into numeric has to be done with care. Just running as.numeric() will not work. See the example below where age is flagged as a factor, perhaps because the variable was stored and exported with the double-quotation marks.
c(100, 101, 102, 103, 104, 105, 106) -> score c("Male", "Female", "Male", "Female", "Female", "Male", "Female") -> sex c("18", "18", "19", "19", "21", "21", "NA") -> age cbind.data.frame(score, sex, age) -> my.dfIf I try as.numeric() the conversion uses the factor codings 1, 2, 3, 4 instead of the actual age values. So what is recommended is that you run as.numeric(levels(data$variable))[data$variable] instead. You can see the difference by comparing age.num1 against age.num2.
as.numeric(my.df$age) -> my.df$age.num1 as.numeric(levels(my.df$age))[my.df$age] -> my.df$age.num2Open up the data-frame and then see these two variables to understand how age.num1 is invalid but age.num2 is valid
We often need to transform an original numeric variable, stored perhaps as a proportion that we want to convert into a percentage. Or we may want to square it, divide it by 100, and so on. These operations are very straightforward.
data.frame(x = c(0.1, 0.2, 0.3, 0.4)) -> my.df # Convert x from proportion to percentmy.df$x * 100 -> my.df$x.pct # Divide x by 10my.df$x / 10 -> my.df$x.div # Square xmy.df$x^2 -> my.df$x.sqrd # Take the square-root of xsqrt(my.df$x) -> my.df$x.sqrt # Multiply x by 2my.df$x * 2 -> my.df$x.dbledThis is the result ...
| x | x% | x/10 | x^2 | x^(1/2) | 2x |
|---|---|---|---|---|---|
| 0.1 | 10 | 0.01 | 0.01 | 0.3162278 | 0.2 |
| 0.2 | 20 | 0.02 | 0.04 | 0.4472136 | 0.4 |
| 0.3 | 30 | 0.03 | 0.09 | 0.5477226 | 0.6 |
| 0.4 | 40 | 0.04 | 0.16 | 0.6324555 | 0.8 |
We can also group numeric data into bins. Let us see this with our reading scores from the hsb2 data-set.
load("data/hsb2.RData")cut(hsb2$read, breaks = c(28, 38, 48, 58, 68, 78)) -> hsb2$grouped_read table(hsb2$grouped_read)## ## (28,38] (38,48] (48,58] (58,68] (68,78] ## 13 69 61 47 9Uh oh! we have a total of 199 so what happened to the 200th? The one that wasn't grouped is a reading score of 28. Why? You have to choose left-open or right-open intervals... should 38 go in the 38-48 group or the 28-38 group? This decision can be invoked by adding right = FALSE or right = TRUE (which is the default). So let me set right = FALSE.
cut(hsb2$read, breaks = c(28, 38, 48, 58, 68, 78), right = FALSE) -> hsb2$grouped_read2 table(hsb2$grouped_read2)## ## [28,38) [38,48) [48,58) [58,68) [68,78) ## 14 68 62 36 20Open hsb2 and arrange rows in ascending order of read. Find read = 48. With the default right = TRUE 48 put in 38-48 group but with right = FALSE 48 put in 48-58 group. So right = FALSE only include values in a group (a,b] if the value is a<=x<b. With right = TRUE only include values in a group [a, b) if the value is a<x<=b.
<div id="htmlwidget-96cbf205c1d7c8d8211d" style="width:100%;height:auto;" class="datatables html-widget"></div><script type="application/json" data-for="htmlwidget-96cbf205c1d7c8d8211d">{"x":{"filter":"none","data":[["1","2","3","4","5","6","7","8","9","10","11","12","13","14","15","16","17","18","19","20","21","22","23","24","25","26","27","28","29","30","31","32","33","34","35","36","37","38","39","40","41","42","43","44","45","46","47","48","49","50","51","52","53","54","55","56","57","58","59","60","61","62","63","64","65","66","67","68","69","70","71","72","73","74","75","76","77","78","79","80","81","82","83","84","85","86","87","88","89","90","91","92","93","94","95","96","97","98","99","100","101","102","103","104","105","106","107","108","109","110","111","112","113","114","115","116","117","118","119","120","121","122","123","124","125","126","127","128","129","130","131","132","133","134","135","136","137","138","139","140","141","142","143","144","145","146","147","148","149","150","151","152","153","154","155","156","157","158","159","160","161","162","163","164","165","166","167","168","169","170","171","172","173","174","175","176","177","178","179","180","181","182","183","184","185","186","187","188","189","190","191","192","193","194","195","196","197","198","199","200"],[57,68,44,63,47,44,50,34,63,57,60,57,73,54,45,42,47,57,68,55,63,63,50,60,37,34,65,47,44,52,42,76,65,42,52,60,68,65,47,39,47,55,52,42,65,55,50,65,47,57,53,39,44,63,73,39,37,42,63,48,50,47,44,34,50,44,60,47,63,50,44,60,73,68,55,47,55,68,31,47,63,36,68,63,55,55,52,34,50,55,52,63,68,39,44,50,71,63,34,63,68,47,47,63,52,55,60,35,47,71,57,44,65,68,73,36,43,73,52,41,60,50,50,47,47,55,50,39,50,34,57,57,68,42,61,76,47,46,39,52,28,42,47,47,52,47,50,44,47,45,47,65,43,47,57,68,52,42,42,66,47,57,47,57,52,44,50,39,57,57,42,47,42,60,44,63,65,39,50,52,60,44,52,55,50,65,52,47,63,50,42,36,50,41,47,55,42,57,55,63],["(48,58]","(58,68]","(38,48]","(58,68]","(38,48]","(38,48]","(48,58]","(28,38]","(58,68]","(48,58]","(58,68]","(48,58]","(68,78]","(48,58]","(38,48]","(38,48]","(38,48]","(48,58]","(58,68]","(48,58]","(58,68]","(58,68]","(48,58]","(58,68]","(28,38]","(28,38]","(58,68]","(38,48]","(38,48]","(48,58]","(38,48]","(68,78]","(58,68]","(38,48]","(48,58]","(58,68]","(58,68]","(58,68]","(38,48]","(38,48]","(38,48]","(48,58]","(48,58]","(38,48]","(58,68]","(48,58]","(48,58]","(58,68]","(38,48]","(48,58]","(48,58]","(38,48]","(38,48]","(58,68]","(68,78]","(38,48]","(28,38]","(38,48]","(58,68]","(38,48]","(48,58]","(38,48]","(38,48]","(28,38]","(48,58]","(38,48]","(58,68]","(38,48]","(58,68]","(48,58]","(38,48]","(58,68]","(68,78]","(58,68]","(48,58]","(38,48]","(48,58]","(58,68]","(28,38]","(38,48]","(58,68]","(28,38]","(58,68]","(58,68]","(48,58]","(48,58]","(48,58]","(28,38]","(48,58]","(48,58]","(48,58]","(58,68]","(58,68]","(38,48]","(38,48]","(48,58]","(68,78]","(58,68]","(28,38]","(58,68]","(58,68]","(38,48]","(38,48]","(58,68]","(48,58]","(48,58]","(58,68]","(28,38]","(38,48]","(68,78]","(48,58]","(38,48]","(58,68]","(58,68]","(68,78]","(28,38]","(38,48]","(68,78]","(48,58]","(38,48]","(58,68]","(48,58]","(48,58]","(38,48]","(38,48]","(48,58]","(48,58]","(38,48]","(48,58]","(28,38]","(48,58]","(48,58]","(58,68]","(38,48]","(58,68]","(68,78]","(38,48]","(38,48]","(38,48]","(48,58]",null,"(38,48]","(38,48]","(38,48]","(48,58]","(38,48]","(48,58]","(38,48]","(38,48]","(38,48]","(38,48]","(58,68]","(38,48]","(38,48]","(48,58]","(58,68]","(48,58]","(38,48]","(38,48]","(58,68]","(38,48]","(48,58]","(38,48]","(48,58]","(48,58]","(38,48]","(48,58]","(38,48]","(48,58]","(48,58]","(38,48]","(38,48]","(38,48]","(58,68]","(38,48]","(58,68]","(58,68]","(38,48]","(48,58]","(48,58]","(58,68]","(38,48]","(48,58]","(48,58]","(48,58]","(58,68]","(48,58]","(38,48]","(58,68]","(48,58]","(38,48]","(28,38]","(48,58]","(38,48]","(38,48]","(48,58]","(38,48]","(48,58]","(48,58]","(58,68]"],["[48,58)","[68,78)","[38,48)","[58,68)","[38,48)","[38,48)","[48,58)","[28,38)","[58,68)","[48,58)","[58,68)","[48,58)","[68,78)","[48,58)","[38,48)","[38,48)","[38,48)","[48,58)","[68,78)","[48,58)","[58,68)","[58,68)","[48,58)","[58,68)","[28,38)","[28,38)","[58,68)","[38,48)","[38,48)","[48,58)","[38,48)","[68,78)","[58,68)","[38,48)","[48,58)","[58,68)","[68,78)","[58,68)","[38,48)","[38,48)","[38,48)","[48,58)","[48,58)","[38,48)","[58,68)","[48,58)","[48,58)","[58,68)","[38,48)","[48,58)","[48,58)","[38,48)","[38,48)","[58,68)","[68,78)","[38,48)","[28,38)","[38,48)","[58,68)","[48,58)","[48,58)","[38,48)","[38,48)","[28,38)","[48,58)","[38,48)","[58,68)","[38,48)","[58,68)","[48,58)","[38,48)","[58,68)","[68,78)","[68,78)","[48,58)","[38,48)","[48,58)","[68,78)","[28,38)","[38,48)","[58,68)","[28,38)","[68,78)","[58,68)","[48,58)","[48,58)","[48,58)","[28,38)","[48,58)","[48,58)","[48,58)","[58,68)","[68,78)","[38,48)","[38,48)","[48,58)","[68,78)","[58,68)","[28,38)","[58,68)","[68,78)","[38,48)","[38,48)","[58,68)","[48,58)","[48,58)","[58,68)","[28,38)","[38,48)","[68,78)","[48,58)","[38,48)","[58,68)","[68,78)","[68,78)","[28,38)","[38,48)","[68,78)","[48,58)","[38,48)","[58,68)","[48,58)","[48,58)","[38,48)","[38,48)","[48,58)","[48,58)","[38,48)","[48,58)","[28,38)","[48,58)","[48,58)","[68,78)","[38,48)","[58,68)","[68,78)","[38,48)","[38,48)","[38,48)","[48,58)","[28,38)","[38,48)","[38,48)","[38,48)","[48,58)","[38,48)","[48,58)","[38,48)","[38,48)","[38,48)","[38,48)","[58,68)","[38,48)","[38,48)","[48,58)","[68,78)","[48,58)","[38,48)","[38,48)","[58,68)","[38,48)","[48,58)","[38,48)","[48,58)","[48,58)","[38,48)","[48,58)","[38,48)","[48,58)","[48,58)","[38,48)","[38,48)","[38,48)","[58,68)","[38,48)","[58,68)","[58,68)","[38,48)","[48,58)","[48,58)","[58,68)","[38,48)","[48,58)","[48,58)","[48,58)","[58,68)","[48,58)","[38,48)","[58,68)","[48,58)","[38,48)","[28,38)","[48,58)","[38,48)","[38,48)","[48,58)","[38,48)","[48,58)","[48,58)","[58,68)"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>read<\/th>\n <th>grouped_read<\/th>\n <th>grouped_read2<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-center","targets":[1,2,3]},{"orderable":false,"targets":0}],"filter":"top","rownames":false,"class":"compact","order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script>More generally you'll see folks just specify the number of cuts they want.
cut(hsb2$read, breaks = 5) -> hsb2$grouped_read3 table(hsb2$grouped_read3)## ## (28,37.6] (37.6,47.2] (47.2,56.8] (56.8,66.4] (66.4,76] ## 14 68 48 50 20But since age cannot include a decimal value this may be unsuitable for our purposes. Of course, we could have avoided this open/closed business of the interval by simply doing this:
cut(hsb2$read, breaks = c(25, 35, 45, 55, 65, 75, 85)) -> hsb2$grouped_read4 table(hsb2$grouped_read4)## ## (25,35] (35,45] (45,55] (55,65] (65,75] (75,85] ## 9 45 76 49 19 2chop() with {santoku}
Say we want the reading scores grouped in 10-point intervals.
library(santoku)chop_width(hsb$read, width = 10) -> hsb$rchop1table(hsb$rchop1)## ## [28, 38) [38, 48) [48, 58) [58, 68) [68, 78) ## 14 68 62 36 20If you want 5, evenly-spaced intervals, which will give each group with a width of exactly 76−285=9.6
chop_evenly(hsb$read, intervals = 5) -> hsb$rchop2table(hsb$rchop2)## ## [28, 37.6) [37.6, 47.2) [47.2, 56.8) [56.8, 66.4) [66.4, 76] ## 14 68 48 50 20What if we want equally spaced cut-points, maybe the quintiles or the quartiles, respectively??
chop_equally(hsb$read, groups = 5) -> hsb$rchop3table(hsb$rchop3)## ## [0%, 20%) [20%, 40%) [40%, 60%) [60%, 80%) [80%, 100%] ## 39 16 62 37 46chop_equally(hsb$read, groups = 4) -> hsb$rchop4table(hsb$rchop4)## ## [0%, 25%) [25%, 50%) [50%, 75%) [75%, 100%] ## 39 44 61 56What if you want specific quantiles?
chop_quantiles(hsb$read, c(0.05, 0.25, 0.50, 0.75, 0.95)) -> hsb$rchop5table(hsb$rchop5)## ## [0%, 5%) [5%, 25%) [25%, 50%) [50%, 75%) [75%, 95%] (95%, 100%] ## 9 30 44 61 47 9Cut-points in terms of the mean and standard deviation?
chop_mean_sd(hsb$read,) -> hsb$rchop6table(hsb$rchop6)## ## [-3 sd, -2 sd) [-2 sd, -1 sd) [-1 sd, 0 sd) [0 sd, 1 sd) [1 sd, 2 sd) [2 sd, 3 sd) ## 2 22 91 39 39 7If you want custom labels ...
chop_quantiles( hsb$read, c(0.05, 0.25, 0.50, 0.75, 0.95), labels = lbl_dash() ) -> hsb$rchop7table(hsb$rchop7)## ## 0% - 5% 5% - 25% 25% - 50% 50% - 75% 75% - 95% 95% - 100% ## 9 30 44 61 47 9chop_quantiles( hsb$read, c(0.05, 0.25, 0.50, 0.75, 0.95), labels = lbl_seq("(A)") ) -> hsb$rchop8table(hsb$rchop8)## ## (A) (B) (C) (D) (E) (F) ## 9 30 44 61 47 9And there you have it!
Changing case: lower/upper/title/sentence/etc
data.frame( sex = c("M", "F", "M", "F"), NAMES = c("Andy", "Jill", "Jack", "Madison"), age = c(24, 48, 72, 96) ) -> my.dfMaybe I want the names of the individuals to be all uppercase, or perhaps all lowercase. I can do this via:
tolower(my.df$sex) -> my.df$sex.lower toupper(my.df$sex) -> my.df$sex.upperMaybe it is not the values but the column names that I want to convert to lowercase. This is easily done via
"names" -> colnames(my.df)[2]Note: I asked for the second column's name to be changed to "names". I could have achieved the same thing by running:
data.frame( sex = c("M", "F", "M", "F"), NAMES = c("Andy", "Jill", "Jack", "Madison"), age = c(24, 48, 72, 96) ) -> my.df tolower(colnames(my.df)) -> colnames(my.df)By the same logic, I could convert each column name to uppercase by typing
toupper(colnames(my.df)) -> colnames(my.df)And then of course, using the following to change a specific column name to uppercase:
data.frame( sex = c("M", "F", "M", "F"), NAMES = c("Andy", "Jill", "Jack", "Madison"), age = c(24, 48, 72, 96) ) -> my.df toupper(colnames(my.df)[1]) -> colnames(my.df)[1] "AGE" -> colnames(my.df)[3]What about title case?
c("somewhere over the rainbow", "to kill a mockingbird", "the reluctant fundamentalist") -> x library(tools)toTitleCase(x) -> x.title x.title## [1] "Somewhere over the Rainbow" "To Kill a Mockingbird" ## [3] "The Reluctant Fundamentalist"When we work with stringi and stringr we'll see other ways of switching letter cases
A little bit about factors
With the hsb data we created some factors. Here I want to show you a couple of things to pay attention to.
First, I can create a new variable and store it as a factor as follows:
my.df$female1[my.df$SEX == "M"] = 1 my.df$female1[my.df$SEX == "F"] = 2 factor( my.df$female1, levels = c(1, 2), labels = c("Male", "Female") ) -> my.df$female1I could have also skipped the 0/1 business and just done this:
my.df$female2[my.df$SEX == "M"] = "Male"my.df$female2[my.df$SEX == "F"] = "Female"factor(my.df$female2) -> my.df$female2Notice the difference between female1 and female2
- female1: Since you specified
levels = c(0, 1)and indicated the lowest level be mapped to Male, the factor is built in that order ... Male first, then Female - female2: Here we did not specify the levels and hence R falls back on its default of building them up alphabetically ... Female first, then Male
Let us see factor levels with a different example. Say we had the following variable, 10 responses to a survey question.
data.frame(x = c( rep("Disagree", 3), rep("Neutral", 2), rep("Agree", 5) ) ) -> fdf factor(fdf$x) -> fdf$responses levels(fdf$responses)## [1] "Agree" "Disagree" "Neutral"table(fdf$responses)## ## Agree Disagree Neutral ## 5 3 2This makes no sense since we'd like the logical ordering of Disagree -> Neutral -> Agree
ordered( fdf$responses, levels = c("Disagree", "Neutral", "Agree") ) -> fdf$newresponses levels(fdf$newresponses)## [1] "Disagree" "Neutral" "Agree"min(fdf$newresponses)## [1] Disagree## Levels: Disagree < Neutral < Agreetable(fdf$newresponses)## ## Disagree Neutral Agree ## 3 2 5I could have also generated this desired order when creating the factor as, for example, via
factor(fdf$x, ordered = TRUE, levels = c("Disagree", "Neutral", "Agree") ) -> fdf$xordered levels(fdf$xordered)## [1] "Disagree" "Neutral" "Agree"min(fdf$xordered)## [1] Disagree## Levels: Disagree < Neutral < Agreetable(fdf$xordered)## ## Disagree Neutral Agree ## 3 2 5Notice the min() command ... which asks for the minimum level of a factor. This works with ordered factors but not with unordered factors; try it with x and with responses.
Before we move on, a word about the command we used to generate a new factor from an existing variable
"Male" -> my.df$female2[my.df$SEX == "M"] "Female" -> my.df$female2[my.df$SEX == "F"]You may run into this a lot when generating new variables, and even when fixing problems with an existing variable. For example, say we were given the data on the sex of these 10 individuals but the values included typos.
data.frame( mf = c( rep("Male", 3), rep("male", 2), rep("Female", 3), rep("femalE", 2) ) ) -> sexdf sexdf$mf = factor(sexdf$mf)levels(sexdf$mf)## [1] "femalE" "Female" "male" "Male"Obviously not what we'd like so we can go in and clean it up a bit. How? As follows:
"Male" -> sexdf$mf[sexdf$mf == "male"] "Female" -> sexdf$mf[sexdf$mf == "femalE"] levels(sexdf$mf)## [1] "femalE" "Female" "male" "Male"table(sexdf$mf)## ## femalE Female male Male ## 0 5 0 5Wait a second, we still see femalE and male but with 0 counts; how should we get rid of these ghosts?
droplevels(sexdf$mf) -> sexdf$newmf levels(sexdf$newmf)## [1] "Female" "Male"table(sexdf$newmf)## ## Female Male ## 5 5Having fixed this issue we can drop the original variable via
NULL -> sexdf$mfSubsetting data and factor levels
What if want only a susbet of the data? For example, say I am working with the diamond data-set and need to subset my analysis to diamonds that are Very Good or better.
library(ggplot2)data(diamonds)str(diamonds$cut)## Ord.factor w/ 5 levels "Fair"<"Good"<..: 5 4 2 4 2 3 3 3 1 3 ...table(diamonds$cut)## ## Fair Good Very Good Premium Ideal ## 1610 4906 12082 13791 21551I can subset as follows:
subset(diamonds, cut != "Fair" & cut != "Good") -> dia.sub1 str(dia.sub1$cut)## Ord.factor w/ 5 levels "Fair"<"Good"<..: 5 4 4 3 3 3 3 5 4 5 ...table(dia.sub1$cut)## ## Fair Good Very Good Premium Ideal ## 0 0 12082 13791 21551Aha! I see no diamonds in the excluded cut ratings but str() still shows R remembering cut as having 5 ratings and the frequency table shows up with zero counts for both so we drop these levels explicitly.
droplevels(dia.sub1$cut) -> dia.sub1$cutnew str(dia.sub1$cutnew)## Ord.factor w/ 3 levels "Very Good"<"Premium"<..: 3 2 2 1 1 1 1 3 2 3 ...table(dia.sub1$cutnew)## ## Very Good Premium Ideal ## 12082 13791 21551Now a bit more on sub-setting data. You can subset with as simple or as complicated a condition you need. For example, maybe you only want Ideal diamonds with a certain minimum clarity and price.
subset(diamonds, cut == "Ideal" & clarity == "VVS1" & price > 3933) -> dia.sub2 table(dia.sub2$cut)## ## Fair Good Very Good Premium Ideal ## 0 0 0 0 283table(dia.sub2$clarity)## ## I1 SI2 SI1 VS2 VS1 VVS2 VVS1 IF ## 0 0 0 0 0 0 283 0min(dia.sub2$price)## [1] 3955Here the sub-setting is generating a data-frame that will only include observations that meet all three requirements (since you used & in the command). If you had run the following instead you would have had very different results.
subset(diamonds, cut == "Ideal" | clarity == "VVS1" | price > 3933) -> dia.sub3 subset(diamonds, cut == "Ideal" & clarity == "VVS1" | price > 3933) -> dia.sub4 subset(diamonds, cut == "Ideal" | clarity == "VVS1" & price > 3933) -> dia.sub5Exporting data
Often, after data have been processed in R, we need to share them with folks who don't use R or want the data in a specific format. This is easily done, as shown below.
data.frame( Person = c("John", "Timothy", "Olivia", "Sebastian", "Serena"), Age = c(22, 24, 18, 24, 35) ) -> out.df write.csv(out.df, file = "data/out.csv", row.names = FALSE) # CSV format library(haven)write_dta(out.df, "out.df.dta") # Stata formatwrite_sav(out.df, "out.df.sav") # SPSS format write_sas(out.df, "data/out.df.sas") # SAS format library(writexl)write_xlsx(out.df, "data/out.df.xlsx") # Excel formatThere are other packages that import/export data so be sure to check them out as well, in particular rio.
Merging data
Say I have data on some students, with test scores in one file and their gender in another file. Each file has a unique student identifier, called ID. How can I create a single data-set? Let us see each data-set first.
data.frame( Score = c(10, 21, 33, 12, 35, 67, 43, 99), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "C31", "Q22") ) -> data1 data.frame( Sex = c("Male", "Female", "Male", "Female", "Male", "Female", "Male", "Female"), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "E52", "H71") ) -> data2Open up both data-sets and note that students C31 and Q22 are missing from data2 and students E52 and H71 are missing from data1.
... here are the two data-sets
<div id="htmlwidget-b171b565011c082c9d51" style="width:100%;height:auto;" class="datatables html-widget"></div><script type="application/json" data-for="htmlwidget-b171b565011c082c9d51">{"x":{"filter":"none","data":[["1","2","3","4","5","6","7","8"],[10,21,33,12,35,67,43,99],["A12","A23","Z14","WX1","Y31","D66","C31","Q22"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>Score<\/th>\n <th>ID<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-center","targets":[1,2]},{"orderable":false,"targets":0}],"filter":"top","rownames":false,"class":"compact","order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script><div id="htmlwidget-bcf4d900b28a98d02dfa" style="width:100%;height:auto;" class="datatables html-widget"></div><script type="application/json" data-for="htmlwidget-bcf4d900b28a98d02dfa">{"x":{"filter":"none","data":[["1","2","3","4","5","6","7","8"],["Male","Female","Male","Female","Male","Female","Male","Female"],["A12","A23","Z14","WX1","Y31","D66","E52","H71"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th> <\/th>\n <th>Sex<\/th>\n <th>ID<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"columnDefs":[{"className":"dt-center","targets":[1,2]},{"orderable":false,"targets":0}],"filter":"top","rownames":false,"class":"compact","order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script>What about the merge? To merge data-frames we need to specify the merge key(s) -- the variable(s) that identifies unique observations in each file. This variable(s) must be present in all the files you want to merge. So long as it exists, you now need to decide: Do you only want to
- merge observations that show up in both files
(natural join) - merge everything even if some cases show up in one but not the other
(full outer join) - merge such that all cases in file
xare retained but only those seen in fileyare joined(left outer join) - merge to keep all cases in
yand only those cases are to be merged fromxthat show up iny, referred to as(right outer join)
merge(x = data1, y = data2, by = c("ID"), all = FALSE) -> natural # only merge if ID seen in both filesmerge(x = data1, y = data2, by = c("ID"), all = TRUE) -> full # merge everything merge(x = data1, y = data2, by = c("ID"), all.x = TRUE) -> left # make sure all cases from file x are retained merge(x = data1, y = data2, by = c("ID"), all.y = TRUE) -> right # make sure all cases from file y are retainedGarrett and Hadley have a wonderful chapter on joins and the diagram below is one of their many descriptors.
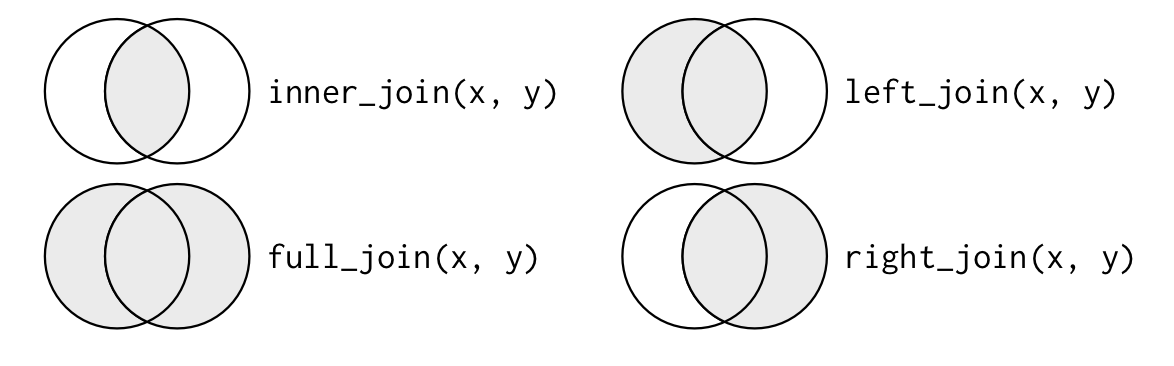
What if the ID variables had different names in the files? You could rename them to have a common name or you could use by.x = and by.y = as shown below.
data.frame( Score = c(10, 21, 33, 12, 35, 67, 43, 99), ID1 = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "C31", "Q22") ) -> data3 data.frame( Sex = c("Male", "Female", "Male", "Female", "Male", "Female", "Male", "Female"), ID2 = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "E52", "H71") ) -> data4 merge( data3, data4, by.x = c("ID1"), by.y = c("ID2"), all = FALSE ) -> diffidsYou can also have more than one merge key. For example, if I am merging data for Ohio schools, each district has a district ID number (dirn), each school has building ID number (birn), and then the district's name (district), the building's name (building), and so on. If I am merging these data then my by = statement will be by = c("dirn", "birn", "district", "building").
If we have multiple data-frames to merge we can do so in several ways but my default approach is to rely on the Reduce() command, as shown below.
data.frame( Score = c(10, 21, 33, 12, 35, 67, 43, 99), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "C31", "Q22") ) -> df1 data.frame( Sex = c("Male", "Female", "Male", "Female", "Male", "Female", "Male", "Female"), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "E52", "H71") ) -> df2 data.frame( Age = c(6, 7, 6, 8, 8, 9, 10, 5), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "E52", "H71") ) -> df3 list(df1, df2, df3) -> my.list Reduce(function(...) merge(..., by = c("ID"), all = TRUE), my.list) -> df.123Here is the merged data-frame ...
| ID | Score | Sex | Age |
|---|---|---|---|
| A12 | 10 | Male | 6 |
| A23 | 21 | Female | 7 |
| C31 | 43 | NA | NA |
| D66 | 67 | Female | 9 |
| E52 | NA | Male | 10 |
| H71 | NA | Female | 5 |
| Q22 | 99 | NA | NA |
| WX1 | 12 | Female | 8 |
| Y31 | 35 | Male | 8 |
| Z14 | 33 | Male | 6 |
Dealing with duplicates
Every now and then you also run into duplicate rows with no good explanation for how that could have happened. Here is an example:
data.frame( Sex = c("Male", "Female", "Male", "Female", "Male", "Female", "Male", "Female"), ID = c("A12", "A23", "Z14", "WX1", "Y31", "D66", "A12", "WX1") ) -> dups.dfStudents A12 and WX1 show up twice! There are two basic commands that we can use to find duplicates -- unique and duplicated.
duplicated(dups.df) # flag duplicate rows with TRUE !duplicated(dups.df) # flag not duplicated rows with TRUE dups.df[duplicated(dups.df), ] # reveal the duplicated rows dups.df[!duplicated(dups.df), ] # reveal the not duplicated rows dups.df[!duplicated(dups.df), ] -> nodups.df # save a dataframe without duplicated rowsdups.df[duplicated(dups.df), ] -> onlydups.df # save a dataframe with ONLY the duplicated rowsRearranging the data-set
We may, at times, want to move the variables around so they are in a particular order (age next to race and so on, for example), perhaps drop some because there are too many variables and we don't need all of them. We may also want to arrange the data-frame in ascending or descending order of some variable(s) (increasing order of age or price for example.
mtcars[order(mtcars$mpg), ] -> df1 # arrange the data-frame in ascending order of mpg mtcars[order(-mtcars$mpg), ] -> df2 # arrange the data-frame in descending order of mpgmtcars[order(mtcars$am, mtcars$mpg), ] -> df3 # arrange the data-frame in ascending order of automatic/manual and then in ascending order of mpgWhat if I want to order the columns such that the first column is am and then comes mpg, followed by qsec and then the rest of the columns? Well, the first thing I want to do is see what variable is in which column via the names() command.
names(mtcars) # the original structure of the mtcars data-frame## [1] "mpg" "cyl" "disp" "hp" "drat" "wt" "qsec" "vs" "am" "gear" "carb"So mpg is in column 1, cyl is in column 2, and carb is the last one in column 11.
Now I'll specify that I want all rows by doing this [, c()] and then specify the order of the columns by entering the column numbers in c()
mtcars[, c(9, 1, 7, 2:6, 8, 10:11)] -> df4 # specifying the position of the columns names(df4)## [1] "am" "mpg" "qsec" "cyl" "disp" "hp" "drat" "wt" "vs" "gear" "carb"If I only wanted certain columns I could simply not list the other columns.
mtcars[, c(9, 1, 7)] -> df5 # specifying the position of the columnsRemember the comma in mtcars[, ... this tells R to keep all rows in mtcars. If, instead, I did this I would only get the first four rows, not all.
mtcars[1:4, c(9, 1, 7)] -> df6 # keep first four rows and the specified columnsI would end up with only the first four rows of mtcars, not every row. So try to remember the order: dataframe[i, j] where i reference the rows and j reference the columns.
Working with strings
Often you run into strings that have to be manipulated. Often, when working with county data, for example, county names are followed by County, Ohio and this is superfluous so we end up excising it
read.csv("data/strings.csv") -> c.data head(c.data)## GEO.id GEO.id2 GEO.display.label respop72016## 1 0500000US39001 39001 Adams County, Ohio 27907## 2 0500000US39003 39003 Allen County, Ohio 103742## 3 0500000US39005 39005 Ashland County, Ohio 53652## 4 0500000US39007 39007 Ashtabula County, Ohio 98231## 5 0500000US39009 39009 Athens County, Ohio 66186## 6 0500000US39011 39011 Auglaize County, Ohio 45894Let us just retain the county name from GEO.display.label
gsub(" County, Ohio", "", c.data$GEO.display.label) -> c.data$county head(c.data[, c(3, 5)])## GEO.display.label county## 1 Adams County, Ohio Adams## 2 Allen County, Ohio Allen## 3 Ashland County, Ohio Ashland## 4 Ashtabula County, Ohio Ashtabula## 5 Athens County, Ohio Athens## 6 Auglaize County, Ohio AuglaizeNote the sequence gsub("look for this pattern", "replace with this pattern", mydata$myvariable)
with and within
If you need to add variables to an existing data frame, you can do it in several ways:
data.frame(x = seq(0, 5, by = 1), y = seq(10, 15, by = 1)) -> dfw dfw$x * dfw$y -> dfw$xy1 # the most basic creation of xy with(dfw, x * y) -> dfw$xy2 # using withwithin(dfw, xy3 <- x * y) -> dfw # using within| x | y | xy1 | xy2 | xy3 |
|---|---|---|---|---|
| 0 | 10 | 0 | 0 | 0 |
| 1 | 11 | 11 | 11 | 11 |
| 2 | 12 | 24 | 24 | 24 |
| 3 | 13 | 39 | 39 | 39 |
| 4 | 14 | 56 | 56 | 56 |
| 5 | 15 | 75 | 75 | 75 |
Notice that if using within() the variable name must be specified as in xy3 <- x * y
ifelse and grepl
The ifelse command is a useful one when creating new variables (invariable factors) from an existing variable
The long way ...
data(mtcars)mtcars$automatic2[mtcars$am == 1] = "Automatic" mtcars$automatic2[mtcars$am == 0] = "Manual"The more efficient way ...
ifelse(mtcars$am == 1, "Automatic", "Manual") -> mtcars$automaticIf you have strings, then grepl comes into play:
load("data/movies.RData")ifelse(grepl("Comedy", movies$genres), "Yes", "No") -> movies$comedy ifelse(grepl("(1995)", movies$title), "Yes", "No") -> movies$in1995 ifelse(grepl("Night", movies$title), "Yes", "No") -> movies$nightIn short, the function looks for the specified string (“Comedy”) in genres and if found adds a flag of “Yes” and if not found adds a flag of “No”
But what if I wanted to run more complicated expressions, for example if the words “Night” or “Dark” or “Cool” are used in the title?
ifelse( grepl("Night", movies$title), "Yes", ifelse(grepl("Dark", movies$title), "Yes", ifelse(grepl("Cool", movies$title), "Yes", "No")) ) -> movies$trickyAh grepl, you beauty!! Nesting our way to what we want ...
grep() and grepl() differ in that both functions find patterns but return different output types
grep()returns numeric values that index the locations where the patterns were foundgrepl()returns a logical vector in which TRUE represents a pattern match and FALSE does not
Run the two commands that follow and focus on the output that scrolls in the console
grep("Comedy", movies$genres)grepl("Comedy", movies$genres)You could use both for sub-setting data as well.
movies[grep("Comedy", movies$genres), ] -> mov1 movies[-grep("Comedy", movies$genres), ] -> mov2Say I only want columns that have the letter “t” in them. No problem:
movies[, grep("t", colnames(movies))] -> mov3 # keep if t is in column name movies[, -grep("t", colnames(movies))] -> mov4 # keep if t is not in column nameTry using grepl instead of grep with the preceding two commands and see what results ...
movies[, grepl("t", colnames(movies))] -> mov3 movies[, -grepl("t", colnames(movies))] -> mov4sums and means
data.frame( x = c(0, 1, 2, 3, 4, NA), y = c(10, 11, 12, NA, 14, 15) ) -> dfs dfs## x y## 1 0 10## 2 1 11## 3 2 12## 4 3 NA## 5 4 14## 6 NA 15rowSums() for each row, sum the columns so that you end up with a row total
rowSums(dfs, na.rm = TRUE) # sum columns 1 and 2 for each row## [1] 10 12 14 3 18 15colSums() for each column, calculate the sum of its values
colSums(dfs, na.rm = TRUE)## x y ## 10 62rowMeans() for each row, calculate the mean of the columns
rowMeans(dfs, na.rm = TRUE)## [1] 5 6 7 3 9 15colMeans() for each column, calculate the mean
colMeans(dfs, na.rm = TRUE)## x y ## 2.0 12.4